Nanosafety Data Interface
Frequently asked questions about eNanoMapper database
Q1.What is an eNanoMapper database instance?
an installation of AMBIT software, implementing the eNanoMapper/AMBIT data model. Usually consists of data from individual nanosafety projects and may be protected by role-based access to keep data confidential.
The database instances offer a user-friendly web interface and an application programming interface (API). They serve as building blocks to feed the aggregated search index and provide interoperability across all or subsets of the instances.
Q2.eNanoMapper/AMBIT data model
a set of data elements and relationships, representing chemical substances with complex composition and associated experimental data, extended to nanomaterials by the EU-funded project eNanoMapper.
More data model details here.
Q3.What is 'PROJECT' - eNanoMapper database (e.g. NANoREG-eNanoMapper database)
a set of eNanoMapper database instances and an aggregated search index used for data management within a project; available at https://enanomapper.adma.ai/projects/{project} ( e.g. for NANoREG database (https://enanomapper.adma.ai/projects/nanoreg)
Q4.The Nanosafety Data Interface
is an online user interface enabling user friendly access to the aggregated search index of (sub)set of eNanoMapper database instances. Usually the user interface is project specific and protected but can be also publicly available.
Multiple project specific interfaces are available at https://enanomapper.adma.ai/
doi:10.1038/s41565-021-00911-6
Q5.Data content
Data from the following projects (
FP7 NANoREG,
H2020 NanoReg2,
eNanoMapper,
H2020 GRACIOUS,
H2020 GRACIOUS (case study pigments),
H2020 GRACIOUS (case study silica),
H2020 caLIBRAte,
H2020 NanoinformaTIX,
H2020 Gov4Nano,
FP7 NanoTest,
FP7 MARINA,
FP7 ENPRA,
NANOGENOTOX,
H2020 RiskGone,
H2020 PATROLS,
H2020 BIORIMA,
FP7 Sanowork,
caNanoLab,
omics metadata,
H2020 SABYDOMA,
H2020 SBD4Nano,
H2020 SAbyNA,
H2020 HARMLESS,
H2020 SUNSHINE,
H2020 CHARISMA,
H2020 CHARISMA,
LRI AMBIT2 read across tool,
H2020 NanoReg2,
H2020 PATROLS,
H2020 POLYRISK,
H2020 PLASTICHEAL,
H2020 PlasticFatE,
H2020 AURORA,
H2020 IMPTOX,
CUSP,
MOMENTUM,
ZEROF,
) was compiled, annotated and imported into separate eNanoMapper database instances.
They can be accessed via the Nanosafety Data Interface at https://enanomapper.adma.ai/ as well as programmatically through REST (API).
Free text and faceted search as well as multiple export and summary reports are available at the search interface (menu Search)
Q6.Data access
The Nanosafety Data Interface at https://enanomapper.adma.ai/ is the main entry point.
Access to protected project areas is granted only to project partners. If you are project partner, contact your WP leader to gain access. Once logged in, an user guide is available at the Help menu.
Programmatic access is available via REST Application Programming Interface (API), subject to the same access restrictions as for the web interface. Contact your WP leader to gain access. Once logged into the web interface, guide for developers is available at the Help menu.
Q7.Data entry
There are several existing approaches to support data entry for regulatory purposes (e.g., OHT), research data in bioinformatics (e.g., ISA-TAB, ISA-JSON)
and its extensions for nanomaterials (e.g., ISA-TAB-Nano),
as well data logging templates originating and widely used in EU funded projects, summarized in the NanoInformatics Roadmap 2030 Chapter 5.
While the eNanoMapper database supports structured formats for data import (e.g. IUCLID5/6 files, W3C RDF), JSON,
the data originating from NanoSafety Cluster during the last decade is predominantly provided in the form of custom spreadsheet (MS Excel) templates.
The eNanoMapper approach has adopted a data provider-friendly workflow which requires only familiar tools and template layouts.
Since ~2017 we have been successful with importing into eNanoMapper databases more than 1500 Excel spreadsheet files.
This is done through a multistage FAIRification workflow using an already defined semantic data model and open source tool (NMDataParser), to map the spreadsheets into the eNanoMapper semantic model and annotate with domain specific ontologies.
Kochev, Nikolay, Nina Jeliazkova, Vesselina Paskaleva, Gergana Tancheva, Luchesar Iliev, Peter Ritchie, and Vedrin Jeliazkov. 2020. “Your Spreadsheets Can Be FAIR: A Tool and FAIRification Workflow for the ENanoMapper Database. Nanomaterials 10 (10): 1908. DOI:10.3390/nano10101908.
To steer the spreadsheet layout design towards harmonization, while retaining the user friendliness and the features deemed important by the labs, we provide a
Template Wizard.
Q8.Template Gallery
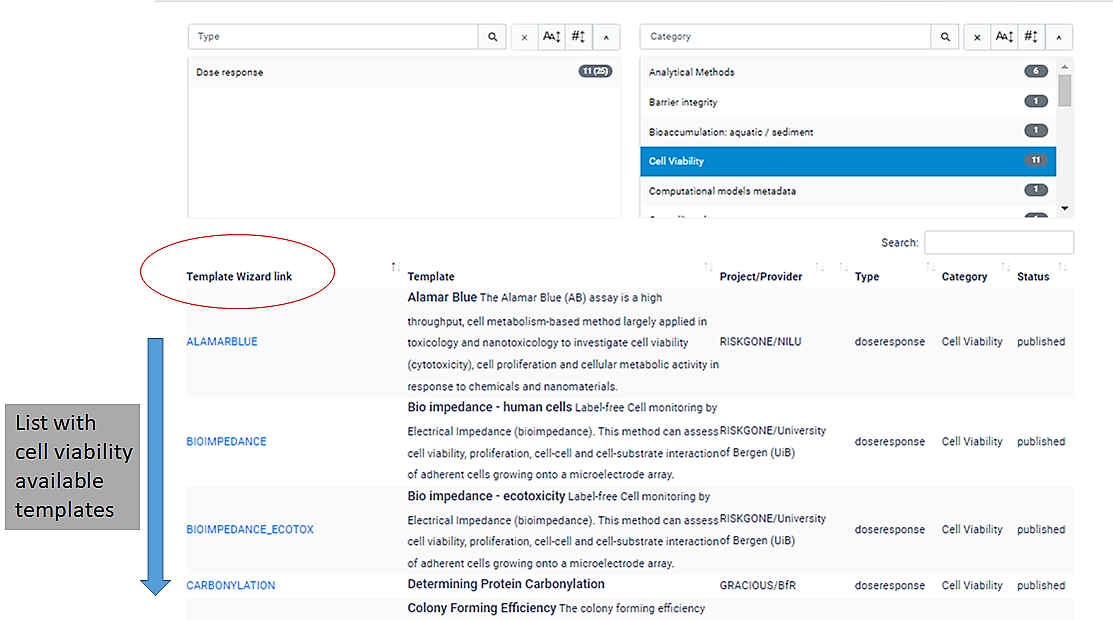
The Template Gallery is an interactive table, in which users can search for tempaltes by type or category.
For example, if you move the slider under the right hand side "Category" search box you will see all of the available templates.
If you click "cell viability", a list with 11 template wizards will be shown.
Click on the name of the specific (cell viability) assay that you have performed. Let's usee the Alamar Blue (AB) assay, for example.
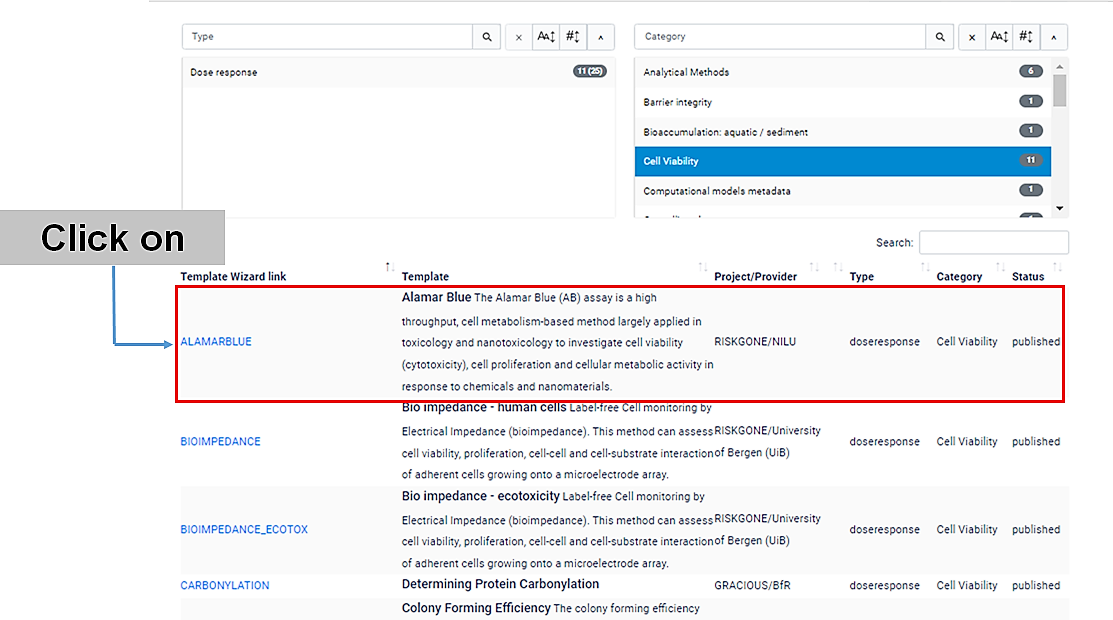 A new window will open , with the Template Wizard.
A new window will open , with the Template Wizard.
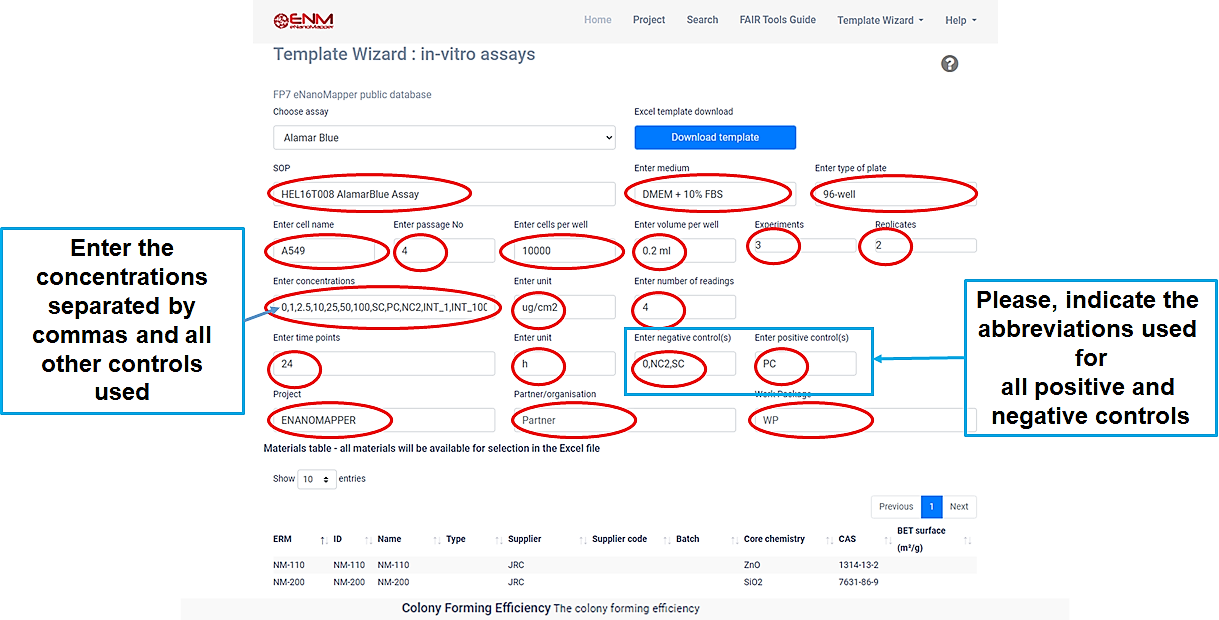
Q9.Template Wizard
The Template Wizard is a web form, where the user can enter metadata of the experiment and download a customized data entry template.
There are different Template Wizards for phychem, ecotox, in-vitro assays and exposure and release experiments.
Before downloading the template, you must fill in all positions marked with red.
Then click on the blue Download template button, and save the file.
- Please consider the user guide (pdf)
- Access the Template Wizard menu for the following project databases:
- The list of of all available template is here.
- The Template Wizard can be extended with templates for different types of assays or different layouts of the existing assays.
Q10.Template styles
The NANoREG approach to the harmonisation of data logging is based on structuring the information on assays in a set of MS Excel templates [DOI:10.2787/505397],
developed by JRC and released under Creative Commons Share-Alike license.
While not strictly following the ISA-TAB and ISA-TAB Nano specifications, the templates have been designed around "ISA-Tab-logic", i.e. structuring the data in investigation-study-assay related groups.
The motivation to create new type of templates instead of using ISA-TAB/ISA-TAB-Nano ones is the perceived low applicability in a more "lab-related" data logging due to lack of user-friendliness.
The NANoREG/JRC templates are being used for data entry in a number of EU funded projects and a subset of the physicochemical templates was refined and published by H2020 Gracious project [DOI:10.2760/142959]
A separate set of Excel templates were developed by the Institute of Occupational Medicine (IOM) and have been used to gather data in several EU FP7 projects (NANOMMUNE, NanoTEST, ENPRA, MARINA, NanoSolutions, SUN) and the COST action MODENA. These (referred as IOM–Nano–EHS style templates) were originally derived in association with the JRC NanoHub from the OECD HT to provide simplified subsets for data collection.
They provide a practical format for end-users collecting the results of physicochemical, in vitro and in vivo toxicology, and more recently eco-toxicology data for a variety of experimental assays addressing nanoEHS. Whilst arranged differently, concentrating on the collection of results, they reflect the principles and include most of the essential metadata features of the ISA-TAB logic, with test method description information and the inclusion of relevant SOPs mandatory requirements
- Haase, & Klaessig. (2018, November 15). EU US Roadmap Nanoinformatics 2030. EU Nanosafety Cluster. DOI:10.5281/zenodo.1486012.
- Kochev, Nikolay, Nina Jeliazkova, Vesselina Paskaleva, Gergana Tancheva, Luchesar Iliev, Peter Ritchie, and Vedrin Jeliazkov. 2020. “Your Spreadsheets Can Be FAIR: A Tool and FAIRification Workflow for the ENanoMapper Database. Nanomaterials 10 (10): 1908. DOI:10.3390/nano10101908.
Besides these, the Template Wizard contains few custom templates (e.g. literature curation, exposure and release templates).
Q11.References
- Nina Jeliazkova et al, 2021. Towards FAIR nanosafety data. Nature Nanotechnology . DOI:10.1038/s41565-021-00911-6.
- Kochev, Nikolay, Nina Jeliazkova, Vesselina Paskaleva, Gergana Tancheva, Luchesar Iliev, Peter Ritchie, and Vedrin Jeliazkov. 2020. “Your Spreadsheets Can Be FAIR: A Tool and FAIRification Workflow for the ENanoMapper Database. Nanomaterials 10 (10): 1908. DOI:10.3390/nano10101908.
- Kochev, Nikolay, Nina Jeliazkova, and Ivanka Tsakovska. 2019. “CHAPTER 3. Chemoinformatics Representation of Chemical Structures – A Milestone for Successful Big Data Modelling in Predictive Toxicology. In Big Data in Predictive Toxicology, edited by Daniel Neagu and Andrea Richarz, 69–107. RSC Publishing. doi:10.1039/9781782623656-00069.
- Jeliazkova, Nina, and Vedrin Jeliazkov. 2019. “CHAPTER 5. Making Big Data Available: Integrating Technologies for Toxicology Applications. In Big Data in Predictive Toxicology (Issues in Toxicology), edited by Daniel Neagu and Andrea-Nicole Richarz, 1 edition, 166–84. Issues in Toxicology. Cambridge: Royal Society of Chemistry. doi:10.1039/9781782623656-00166.
- Jeliazkova, N., V. Koch, Q. Li, U. Jensch, J. Schneider Reigl, R. Kreiling, I. Georgiev, and B. Hubesch. 2016. “Linking LRI AMBIT Chemoinformatic System with the IUCLID Substance Database to Support Read-across of Substance Endpoint Data and Category Formation. Toxicology Letters 258 (September): S114–15. doi:10.1016/j.toxlet.2016.06.1469
- Jeliazkova, Nina, Charalampos Chomenidis, Philip Doganis, Bengt Fadeel, Roland Grafström, Barry Hardy, Janna Hastings, et al. 2015. “The eNanoMapper Database for Nanomaterial Safety Information. Beilstein Journal of Nanotechnology 6 (July): 1609 34. doi:10.3762/bjnano.6.165.
- Hastings, Janna, Nina Jeliazkova, Gareth Owen, Georgia Tsiliki, Cristian R Munteanu, Christoph Steinbeck, and Egon Willighagen. 2015. “eNanoMapper: Harnessing Ontologies to Enable Data Integration for Nanomaterial Risk Assessment. Journal of Biomedical Semantics 6 (1): 10. doi:10.1186/s13326-015-0005-5.
- Jeliazkova, Nina, Philip Doganis, Bengt Fadeel, Roland Grafstrom, Janna Hastings, Vedrin Jeliazkov, Pekka Kohonen, et al. 2014. “ Prototype: A Substance Database to Support Safe-by-Design. In 2014 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 1–9. IEEE. doi:10.1109/BIBM.2014.6999367.
- Jeliazkova, Nina, and Vedrin Jeliazkov. 2011. “AMBIT RESTful Web Services: An Implementation of the OpenTox Application Programming Interface. Journal of Cheminformatics 3 (1): 18. doi:10.1186/1758-2946-3-18
